Plotting NCAR MUSICA outputs using R
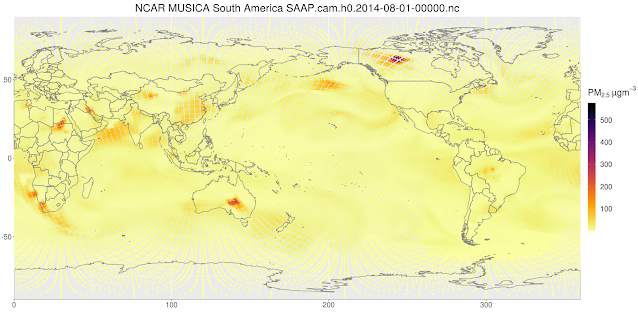
library(ncdf4) library(colorout) library(ggplot2) library(cptcity) fc <- list.files(path = "/glade/scratch/sibarra/SAAP/run", pattern = "SAAP.cam.h0.2014", full.names = T) nc <- list.files(path = "/glade/scratch/sibarra/SAAP/run", pattern = "SAAP.cam.h0.2014") dates <- as.Date(nc, format = "SAAP.cam.h0.%Y-%m-%d-00000.nc") nd <- paste0(strftime(dates, "%Y%m%d"), ".nc") mi <- map_data('world', wrap=c(0,360)) # geo #### lat <- ncvar_get(nc1, varid = "lat") lon <- ncvar_get(nc1, varid = "lon") #PM25 #### x <- ncvar_get(nc1, varid = "PM25") dtx <- data.table::data.table( lat = lat, lon = lon, x = x[, 32]*1e9 ) summary(dtx) rc <- classInt::classIntervals(var = dtx$x, n = 100, style = "quantile")$brks ggplot(dtx, aes(x